import numpy as np
import scipy.spatial.distance as dist
import scipy.linalg as linalg
import matplotlib.pyplot as plt
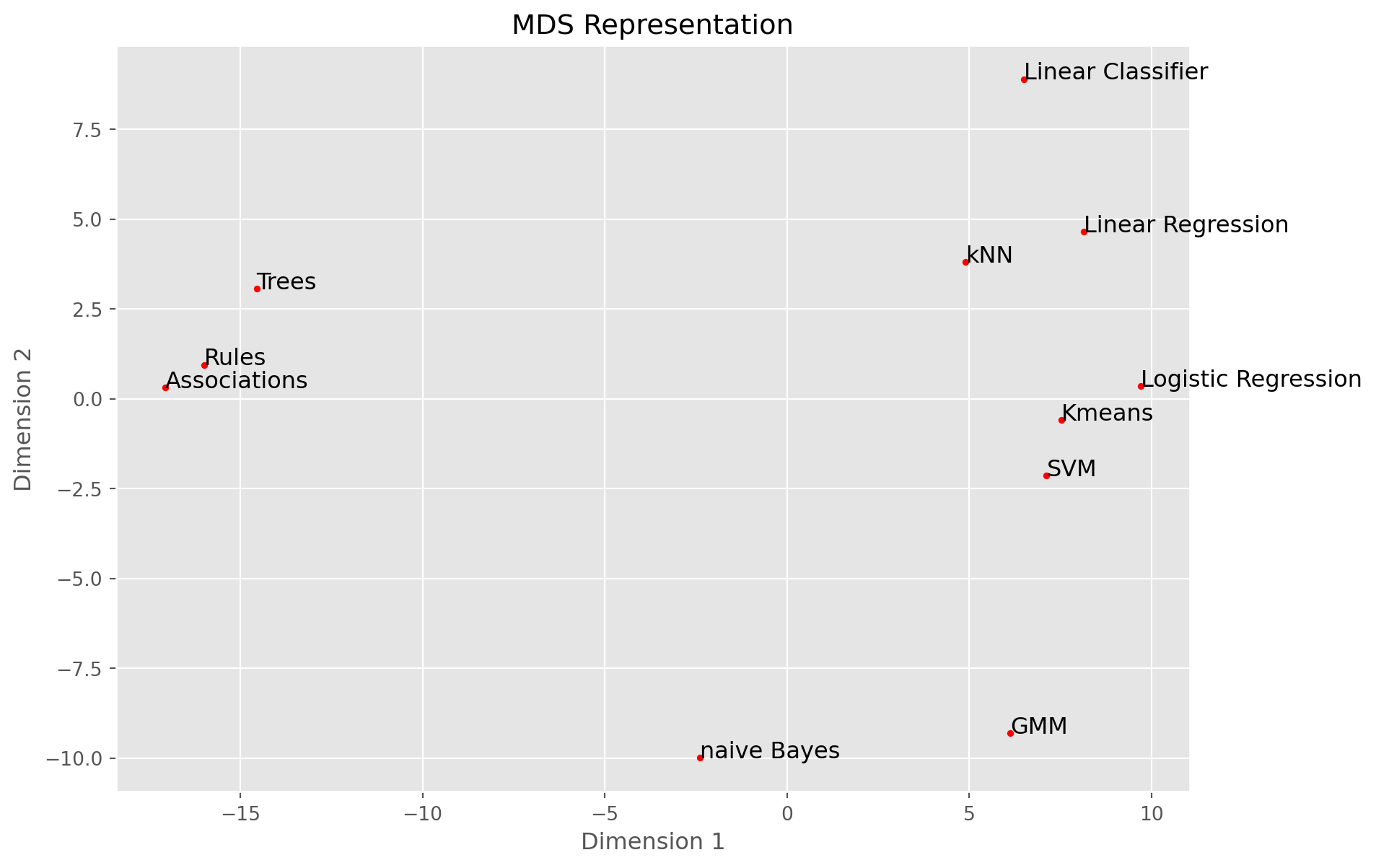
names = ['Trees', 'Rules', 'naive Bayes', 'kNN', 'Linear Classifier', 'Linear Regression',
'Logistic Regression', 'SVM', 'Kmeans', 'GMM', 'Associations']
features = ['geom', 'stat', 'logic', 'group', 'grad', 'symb', 'real', 'sup', 'unsup', 'multi']
M = np.array([
[1,0,3,3,0,3,2,3,2,3],
[0,0,3,3,1,3,2,3,0,2],
[1,3,1,3,1,3,1,3,0,3],
[3,1,0,2,2,1,3,3,0,3],
[3,0,0,0,3,1,3,3,0,0],
[3,1,0,0,3,0,3,3,0,1],
[3,2,0,0,3,1,3,3,0,0],
[2,2,0,0,3,2,3,3,0,0],
[3,2,0,1,2,1,3,0,3,1],
[1,3,0,0,3,1,3,0,3,1],
[0,0,3,3,0,3,1,0,3,1]
])
plt.style.use('ggplot')
w1, w2, w3 = 5, 3, 1
W = np.array([w1, w1, w1, w2, w2, w3, w3, w3, w3, w3])
M = M * W
D = dist.pdist(M, metric='euclidean')
D = dist.squareform(D)
def cmdscale(D):
n = D.shape[0]
H = np.eye(n) - np.ones((n, n)) / n
B = -0.5 * H @ (D ** 2) @ H
eigvals, eigvecs = linalg.eigh(B)
idx = np.argsort(eigvals)[::-1]
eigvals = eigvals[idx]
eigvecs = eigvecs[:, idx]
return eigvecs[:, :2] * np.sqrt(eigvals[:2]), eigvals
Y, eigvals = cmdscale(D)
plt.figure(figsize=(10,7))
plt.scatter(Y[:, 0], Y[:, 1], c='r', marker='.')
for i, name in enumerate(names):
plt.text(Y[i, 0], Y[i, 1], name, fontsize=12)
plt.xlabel('Dimension 1')
plt.ylabel('Dimension 2')
plt.title('MDS Representation')
plt.show()